ROBIN: A Complete AI-Driven Scientific Discovery System
Executive Summary
Researchers at FutureHouse have developed ROBIN, a multi-agent AI system capable of fully automating the entire scientific discovery process—from literature review through hypothesis generation, experimental design, data analysis, and iterative refinement. The system successfully identified ripasudil as a novel therapeutic candidate for dry age-related macular degeneration (dAMD), demonstrating the potential for autonomous AI-driven drug discovery.
Breaking the Scientific Discovery Bottleneck
The traditional scientific process involves a complex iterative cycle: researchers conduct background literature reviews, generate hypotheses, design experiments, analyze results, and refine their understanding based on findings. While recent AI advances have tackled individual components of this workflow, no system has previously integrated all these steps into a single, autonomous platform capable of driving genuine scientific discovery.
This limitation is particularly acute in therapeutic development, where the synthesis of vast literature across multiple domains creates a significant bottleneck. Drug repurposing exemplifies this challenge—countless therapeutic opportunities likely exist within existing scientific literature, but the cognitive load required to connect disparate insights across biological, clinical, and pharmaceutical knowledge domains often delays discovery by years or decades.
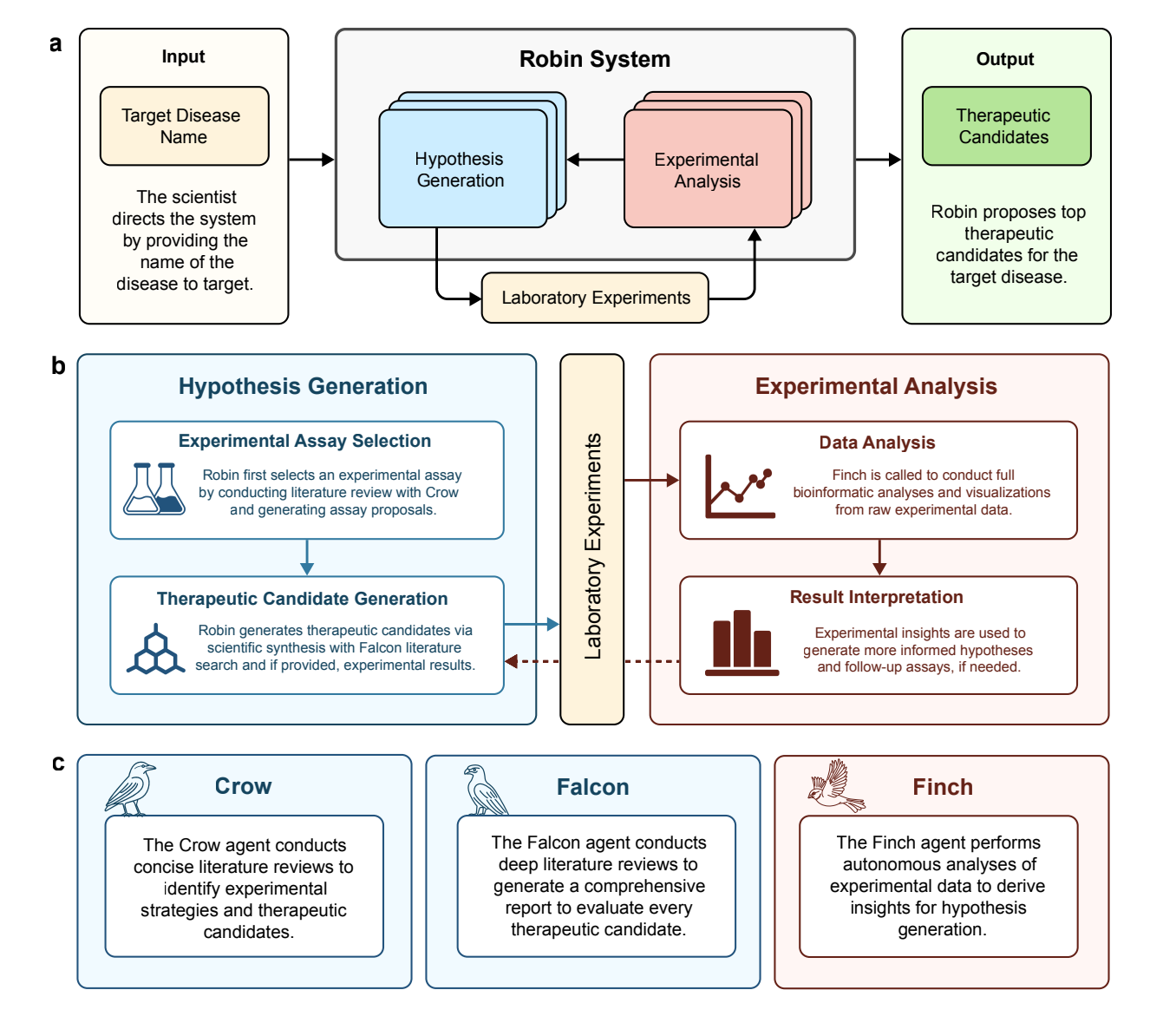
Multi-Agent Architecture: Modeling the Scientific Method
ROBIN represents a fundamental architectural advancement through its implementation of specialized agents that mirror distinct cognitive processes in scientific reasoning:
Crow Agent: Conducts concise, targeted literature searches using PaperQA2, which achieves expert-level performance in information retrieval across scientific literature, clinical trials, and databases like Open Targets Platform.
Falcon Agent: Performs comprehensive deep literature reviews to generate detailed evaluations of therapeutic candidates, providing both scientific rationale and potential limitations.
Finch Agent: Executes autonomous scientific data analysis across multiple experimental modalities including RNA-seq, flow cytometry, and other bioinformatics workflows using standardized Docker environments.
The system orchestrates these agents through structured workflows that automate hypothesis generation, experimental strategy selection, and iterative refinement based on experimental results. Critically, ROBIN employs an LLM-judged tournament system using the Bradley-Terry-Luce model to rank hypotheses and experimental strategies, with demonstrated alignment to human expert preferences.
Technical Implementation: Lab-in-the-Loop Discovery
ROBIN's experimental workflow demonstrates several key technical innovations:
Consensus-Driven Analysis: When analyzing experimental data, ROBIN launches multiple parallel Finch trajectories (typically 10) that independently process the same dataset. This approach leverages the stochasticity of language agents to explore diverse analytical approaches while achieving consensus-driven conclusions that prove more reliable than single-trajectory analyses.
Automated Tournament Ranking: The system uses pairwise comparisons adjudicated by Claude 3.7 Sonnet to rank up to 30 therapeutic candidates. For larger hypothesis sets, 300 random pairwise comparisons provide comprehensive assessment within computational constraints.
Iterative Experimental Design: Unlike static prediction systems, ROBIN actively proposes follow-up experiments based on initial results, enabling true iterative discovery cycles that mirror human scientific reasoning.
Human-AI Collaboration Framework: The system maintains a "scientist-in-the-loop" paradigm where researchers execute physical experiments while ROBIN handles intellectual synthesis and analysis tasks.
Figure 1: Architecture and workflow of the Robin system.
Real-World Validation: Autonomous Discovery of Ripasudil for dAMD
The system's capabilities were validated through application to dry age-related macular degeneration, a leading cause of blindness affecting 1.5 million Americans with limited treatment options. ROBIN's discovery process proceeded as follows:
Literature Synthesis: ROBIN analyzed 151 papers to propose ten biologically relevant dAMD mechanisms, ultimately selecting enhanced RPE cell phagocytosis as the optimal therapeutic strategy.
Candidate Generation: The system reviewed approximately 400 papers on RPE phagocytosis and proposed 30 therapeutic candidates, ranking them through comprehensive Falcon evaluations.
Experimental Validation: Initial testing of five top candidates identified Y-27632 (a ROCK inhibitor) as significantly enhancing RPE phagocytosis. Subsequent RNA-seq analysis revealed 3-fold upregulation of ABCA1, a critical lipid efflux pump implicated in macular degeneration pathogenesis.
Iterative Refinement: ROBIN's second iteration proposed ripasudil, a clinically-approved ROCK inhibitor for glaucoma treatment in Japan. Experimental validation showed ripasudil achieved 7.5-fold enhancement of RPE phagocytosis compared to controls, significantly outperforming Y-27632.
Technical Limitations and Research Frontiers
Several important technical challenges remain for fully autonomous discovery systems:
Experimental Protocol Generation: While ROBIN generates experimental outlines, it cannot yet produce detailed, executable laboratory protocols without human interpretation.
Prompt Engineering Dependencies: Finch's analytical reliability currently requires domain expert prompt engineering for specific data modalities, limiting truly autonomous operation.
Evaluation Alignment: The LLM-judged tournament system, while demonstrating good concordance with human experts (7.25/10 overlap in top hypotheses), may benefit from improved alignment with scientific judgment criteria.
Reproducibility Across Domains: Validation has focused primarily on therapeutic discovery; broader applicability across diverse scientific domains requires further investigation.
Future Directions: Towards Autonomous Scientific Intelligence
The success of ROBIN's integrated approach suggests several promising technical developments:
Closed-Loop Experimentation: Integration with automated laboratory systems could enable fully autonomous experimental cycles without human intervention for routine assays.
Multimodal Data Integration: Expanding beyond text-based literature to incorporate experimental databases, protein structures, and chemical libraries could enhance hypothesis quality.
Cross-Domain Discovery: Applying similar architectures to fundamental research questions beyond therapeutics could accelerate discovery across scientific disciplines.
Collaborative AI Networks: Enabling multiple ROBIN instances to share insights and coordinate research efforts could model large-scale scientific collaboration.
Questions for Further Reflection:
As Human AI co-intelligence (HAIXBIO) systems become reality, the scientific community must continue addressing:
Attribution and Credit: How should attribution and credit be managed in discoveries where AI systems contribute significantly to hypothesis generation and experimental design?
Reproducibility and reliability: What safeguards and validation frameworks will be necessary to ensure the reliability, reproducibility and safety of therapeutics discovered through autonomous AI systems?
Speed: How might the integration of real-time experimental feedback change the fundamental pace of scientific discovery across disciplines?
Conclusions: Implications for Biological and Drug Discovery
Biology impact: Novel Mechanistic Insights
ROBIN's mechanistic insights demonstrate how AI-driven discovery can reveal novel molecular connections within disease pathways. ROBIN analysis revealed previously unexplored connections between ROCK inhibition and lipid metabolism in RPE cells. The discovery of ABCA1 upregulation upon ROCK inhibitor treatment suggests a novel therapeutic mechanism where enhanced phagocytosis couples with improved lipid efflux—both critical functions that deteriorate in dAMD pathogenesis. This finding connects to broader macular degeneration biology: ABCA1 belongs to the same transporter family as ABCA4, a known therapeutic target, while its lipid acceptor Apo-E has also been identified as a potential dAMD target.
Industry Impact: Transforming Drug Discovery Economics
The drug repurposing focus proves particularly valuable given the extensive lag times between scientific insights and therapeutic applications in orphan diseases and small indications. ROBIN's ability to automate literature synthesis, hypothesis generation and experimental strategy positions it to identify previously overlooked connections between established compounds and novel therapeutic opportunities. ROBIN could dramatically accelerate early-stage discovery in orphan diseases and indication expansion strategies.