The Virtual Lab Revolution
How AI Agents Conducted Real Interdisciplinary Science and Designed Functional Nanobodies
Executive Summary
Researchers at Stanford and the Chan Zuckerberg Biohub have created a groundbreaking "Virtual Lab" - a multi-agent AI system where specialized AI researchers collaborate through structured meetings to conduct sophisticated interdisciplinary science. Their proof-of-concept designed 92 SARS-CoV-2 nanobodies with over 90% experimental success rates, demonstrating AI's potential as a true research partner rather than mere tool.
Breaking the Interdisciplinary Barrier
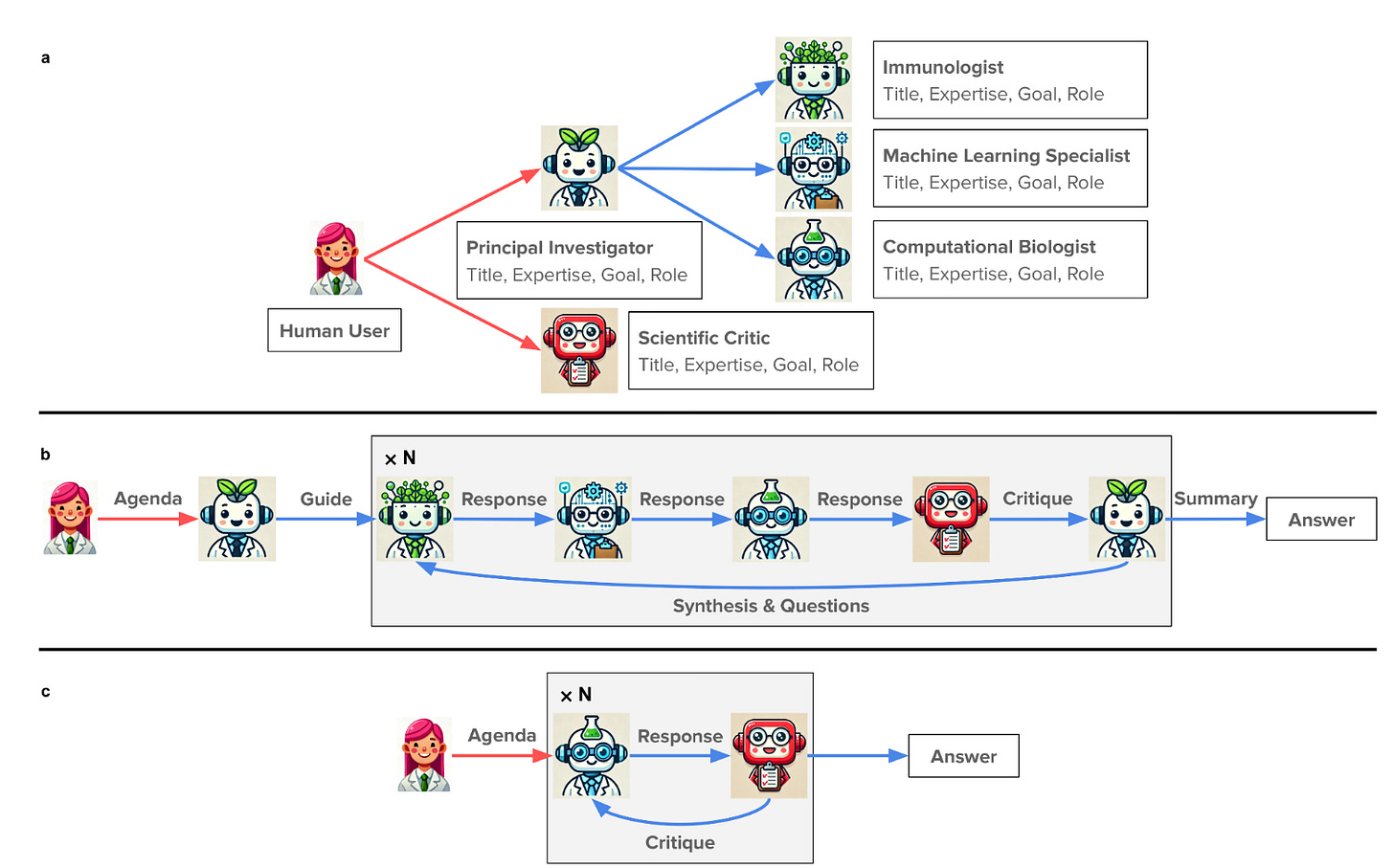
Modern scientific breakthroughs increasingly require interdisciplinary collaboration, yet building and coordinating teams across fields remains challenging. The Virtual Lab addresses this by creating AI agents with distinct scientific expertise - immunologists, computational biologists, machine learning specialists - that collaborate through structured meetings guided by a Principal Investigator agent.
Unlike other AI-for-science approaches that treat AI as a tool, the Virtual Lab represents a paradigm shift toward AI as a collaborative research partner. The system doesn't just answer questions or run calculations; it participates in the entire research process from hypothesis generation to experimental design.
Multi-Agent Architecture: Computational Scientific Collaboration
The Virtual Lab implements several innovative architectural components:
Specialized Agent Roles:
Principal Investigator Agent: Leads meetings, synthesizes discussions, makes strategic decisions
Scientist Agents: Domain experts (immunologist, computational biologist, ML specialist) with specific expertise, goals, and roles
Scientific Critic Agent: Provides critical feedback and identifies errors across all interactions
Meeting Framework:
Team Meetings: All agents discuss broad research directions collaboratively
Individual Meetings: Single agents tackle specific technical implementations with critic feedback
Parallel Meetings: Multiple versions run simultaneously, then merged for optimal outcomes
This structure mirrors human research collaboration while leveraging AI's ability to process vast information and maintain consistency across complex discussions.
Virtual Lab architecture
Technical Innovation: ESM + AlphaFold + Rosetta Pipeline
The Virtual Lab designed a sophisticated computational workflow combining three state-of-the-art tools:
ESM (Evolutionary Scale Modeling): Protein language model calculating log-likelihood ratios for mutations
AlphaFold-Multimer: Predicting nanobody-spike protein complex structures
Rosetta: Computing binding energies and structure refinement
A weighted score balances evolutionary likelihood, structural confidence, and binding affinity - a sophisticated approach that required reasoning across multiple scientific domains.
Notably, the selection of these tools were decided by the PI agent running parallel meetings with the machine learning and computational biologist agent. Python scripts to use the selected tools were coded in python by the machine learning and computational biologist agents.
Experimental Validation: From Computation to Bench
The true test comes in the lab. Of 92 designed nanobodies:
90%+ expressed and remained soluble (35/92 showed high expression)
Two promising candidates emerged:
Nb21 mutant (I77V-L59E-Q87A-R37Q): Gained binding to JN.1 and KP.3 variants
Ty1 mutant (V32F-G59D-N54S-F32S): Improved Wuhan binding, gained JN.1 binding
These results suggests the Virtual Lab's ability to design functional biomolecules, not just propose theoretical improvements.
Technical Analysis: What Made This Work?
Iterative Optimization: Four rounds of mutation with top-5 selection at each stage enabled gradual improvement while maintaining diversity.
Multi-Metric Evaluation: Combining evolutionary fitness (ESM), structural confidence (AlphaFold), and binding energy (Rosetta) provided robust candidate selection.
Human-AI Balance: Humans provided ~1.3% input (high-level guidance, agenda setting) while AI handled detailed implementation and reasoning. Humans in the loop also provide specific instructions on how to modify python code to run the tools and run the workflow. Feedback was provided through detailed prompts
Parallel Processing + Merging: Running multiple meeting variants and intelligently combining outputs improved consistency and quality over single-shot approaches.
Biological Implications: Beyond Nanobody Design
The Virtual Lab's success suggests potential broad applications:
Drug Discovery: Accelerated lead optimization and target identification
Protein Engineering: Rational design of enzymes and therapeutic proteins
Biomarker Discovery: Hypothesis generation for diagnostic targets
Systems Biology: Integration of multi-omics data for pathway elucidation
Limitations and Future Directions
Current Constraints:
Knowledge cutoff limitations of the LLM used for the agents (it missed latest tools like AlphaFold 3)
Requires prompt engineering for optimal performance
“Limited” to problems with computable evaluation metrics. Computable evaluation metrics is a must when evaluating and testing predictions from AI Agentic systems for scientific research
Future Opportunities:
Integration with emerging Agentic standard protocols: MCP, A2A and other emerging ones.
Grounding and Real-time literature integration via RAG and web search (Included in other systems such as google’s AI Co-Scientist)
Closed-loop laboratory automation. As suggested by emerging “lab-in-loop” approaches
Multi-institutional collaborative research networks
Domain-specific fine-tuning for specialized fields
Industry Impact: The Research Acceleration Paradigm
The Virtual Lab represents more than incremental improvement - it's a new research methodology. Organizations implementing similar systems could:
Compress discovery timelines from months to days
Access interdisciplinary expertise without hiring specialists
Generate higher-quality hypotheses through Human-AI co-intelligence (HAIXBIO)
Focus human scientists on creatively tackling larger problems and hypotheses, derive insights and define critical experimental validation
Critical Questions for the Field
As Human AI co-intelligence (HAIXBIO) systems become reality, the scientific community must address:
Attribution and Credit: How do we describe, and transparently acknowledge AI contributions to scientific discovery?
Reproducibility: Developing frameworks and benchmarks to independently verify Human AI co-intelligence systems generated hypotheses
Bias and Diversity: How do we ensure AI doesn't narrow scientific thinking?
Education: How should we train the next generation of scientist and retrain current scientist adopting AI-collaborative systems?
Ethics: What guardrails are needed for Human AI co-intelligence driven scientific research?
Conclusions: Toward Human-AI Research Symbiosis
The Virtual Lab suggests that AI can be more than a sophisticated tool - it will become a genuine research collaborator in the near future. By combining multiple specialized agents with structured interaction protocols, the system achieved results that would have required months of traditional research.
This, and similar works reviewed here in HAixBio™ news, opens the door to a future where human creativity and intuition are combined with AI's computational power to accelerate scientific discovery. The question will not longer be whether AI will transform research, but how quickly we can develop frameworks for productive Human AI co-intelligence and collaboration.
For data scientists and AI researchers, the Virtual Lab provides a template for building multi-agent systems that can tackle complex scientific challenges. The key insight, true Human AI science collaboration requires systems that can engage in the full scientific process in a tight feed-back loop: hypothesis creation, (large scale) data generation, hypothesis testing and validation; not just execute individual tasks.